Task Overview
Task 3 addresses the critical challenge of cross-context generalization. Models trained on perturbation data from one cell type or experimental condition must predict responses in completely different cellular contexts.
Key Challenges:
- Cell Type Specificity: Different cell types show distinct perturbation responses
- Context Adaptation: Experimental conditions and protocols vary across datasets
- Domain Transfer: Bridging gaps between different experimental platforms
- Biological Variability: Accounting for inherent biological differences
Transfer Scenarios:
- Inter-cell type: From one cell line to another
- Inter-dataset: Across different experimental studies
- Inter-condition: Different culture or treatment conditions
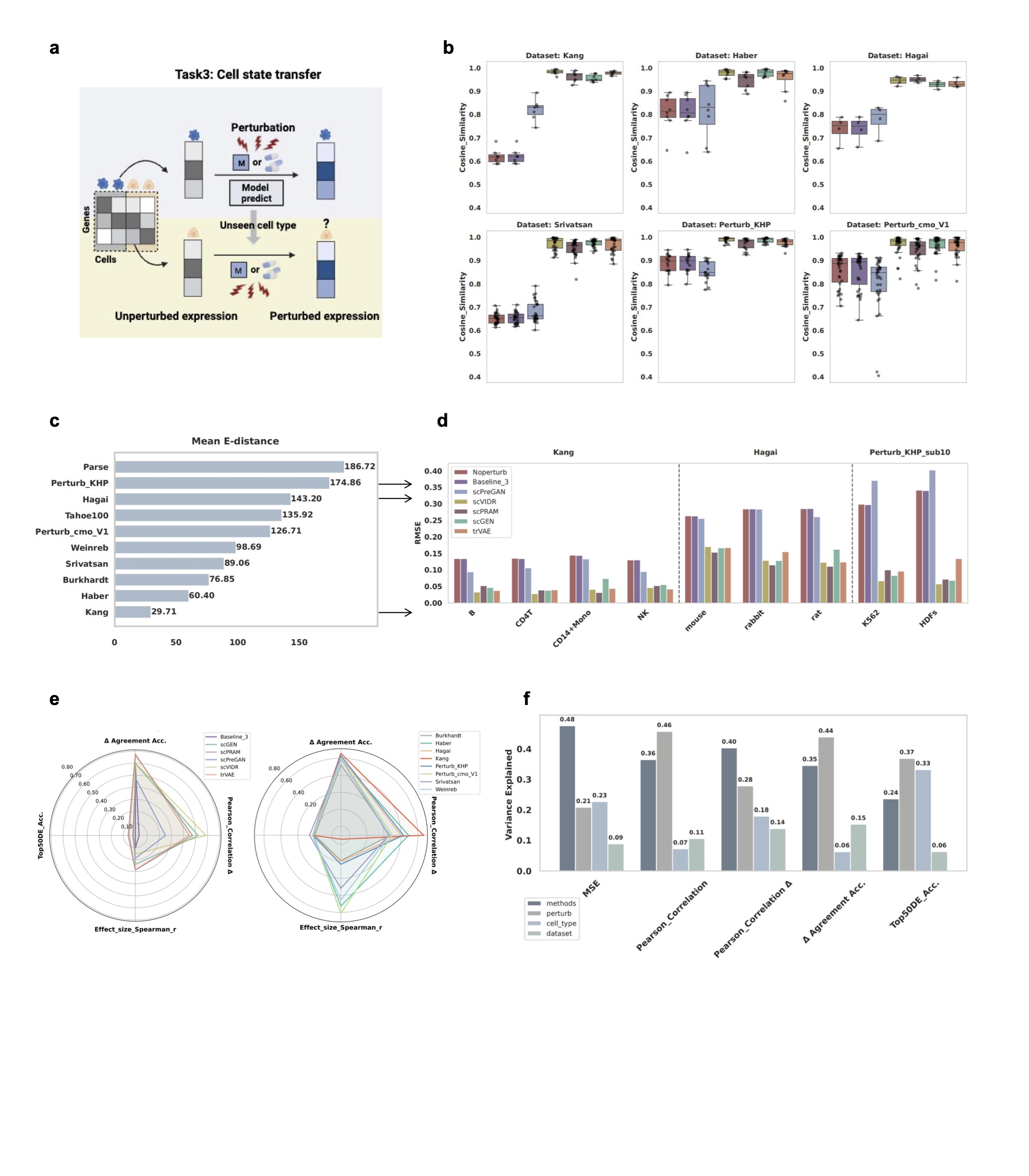
Figure 4: Task 3 results showing cell state transfer performance across different datasets and contexts