Task Overview
In Task 1, models are trained on a subset of perturbations and tested on their ability to predict responses to completely unseen perturbations. This represents a critical real-world scenario where researchers need to predict the effects of new drugs or genetic modifications.
Key Challenges:
- Generalization: Models must learn transferable patterns from seen perturbations
- Effect Size Dependency: Performance varies significantly with perturbation magnitude
- Cell Type Specificity: Responses can be highly context-dependent
Evaluation Metrics:
- Pearson Correlation
- R-squared
- Mean Squared Error (MSE)
- Mean Absolute Error (MAE)
- Cosine Similarity
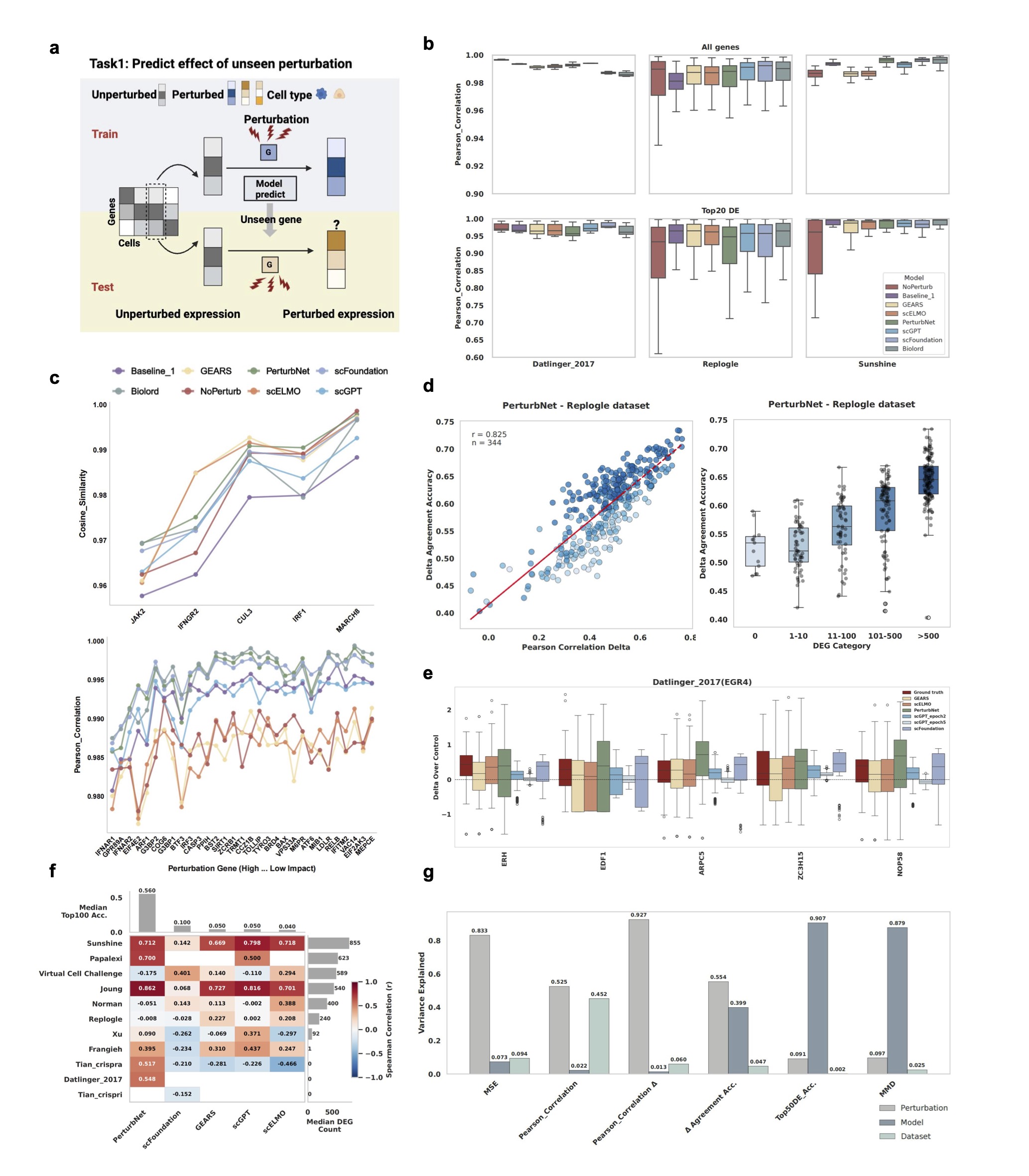
Figure 2: Task 1 performance results showing model comparison across different metrics and effect sizes